TetraRNADB 
A database include the landscape of human tetra-classification RNAs
RNAs are usually divided into protein-coding RNAs and non-coding RNAs based on their protein coding potential, and believed to function as either protein or RNA itself. However, Increasing researches suggested that the function for RNA maybe not unitary, but could be bilateral. These RNAs with both coding and non-coding functions are called cncRNAs or bi-functional RNAs [1-5].
Here we extract the Inherent nucleotide sequence features, Inherent peptide features, structural features and expert features from known human mRNAs, lncRNAs and two kind of bi-functional RNAs t o build a tetra-classification model via random forest algorithm (TetraRNA model), and finally get the landscape of human tetra-classification RNAs.
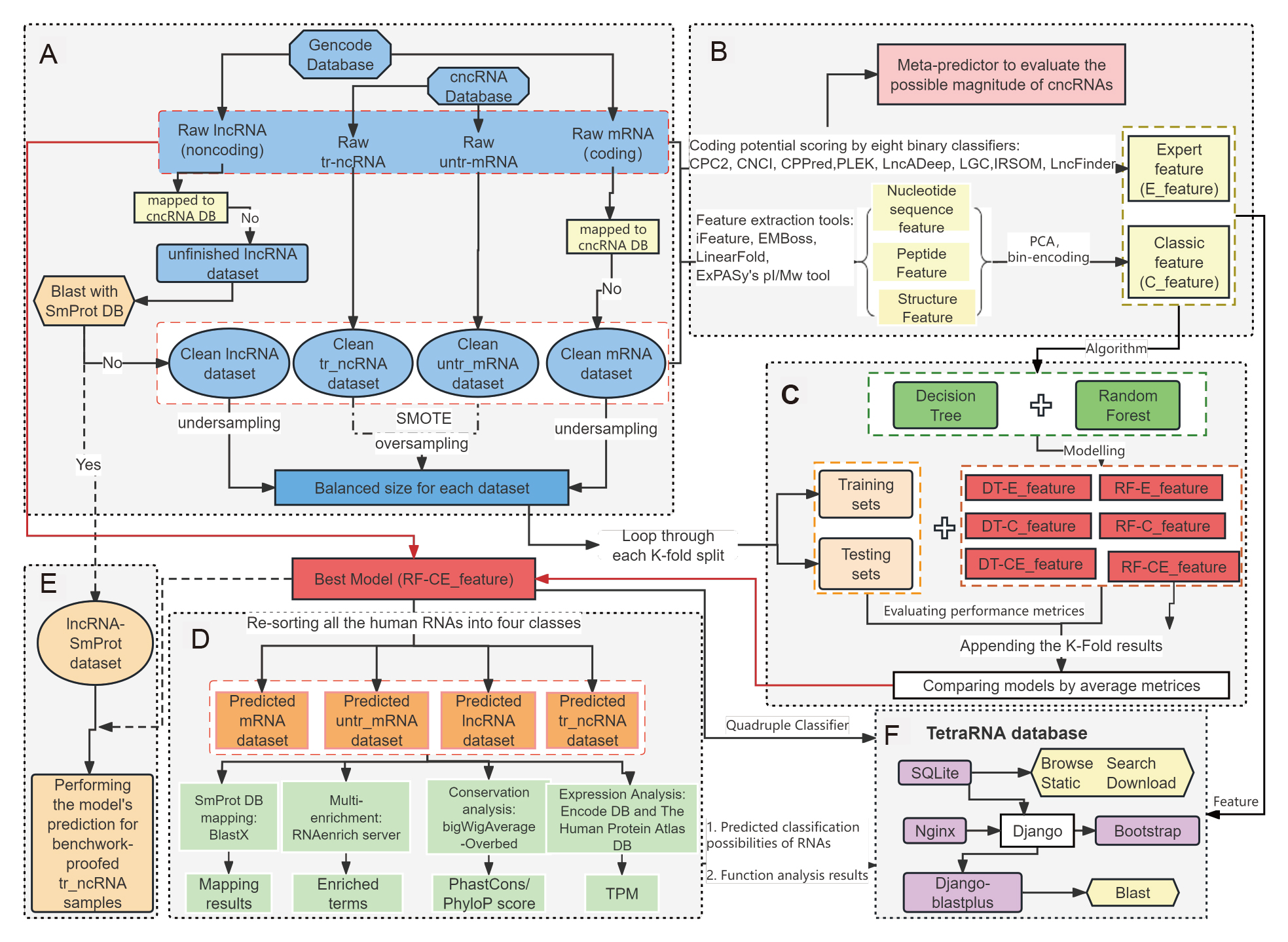
According to our TetraRNA model, human RNAs are divided into four categories: protein-coding RNAs (mRNAs), long noncoding RNAs (lncRNAs), bi-functional RNAs in which non-coding is primary and coding is accessorial (translated non-coding RNAs, tr-ncRNAs), and bi-functional RNAs in which coding is primary and non-coding is accessorial (untranslated mRNAs, untr-mRNAs). We finally have 83729 mRNAs, 33425 lncRNAs, 23235 tr-ncRNAs and 951 untr-mRNAs.
TetraRNA database provide the tetra-classification probabilities for human RNAs based on our model. This database displays the original types and new tetra types of human RNAs and also shows the primary analyses of human four classification RNAs.



